
Among the complex circuitry of AML signaling systems, the menin pathway is thought to have a far-reaching impact on AML development and outcomes.1-7
The menin pathway is a crucial mediator in gene expression driving leukemogenesis1,2,4,8

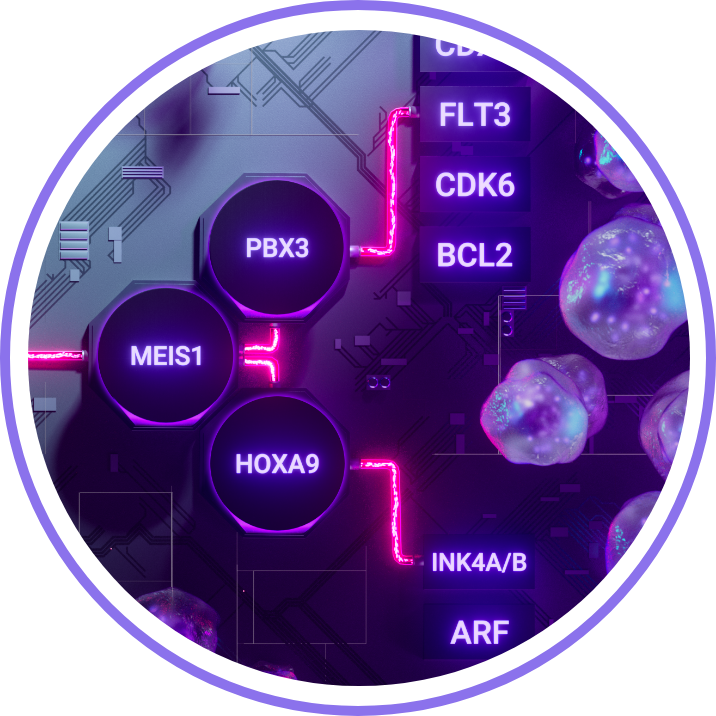
By decoding the mechanisms through which menin affects leukemic cell behavior, it is possible to enhance the understanding of menin-dependent AMLs2,4